I focus on genetic programming in reinforcement learning tasks. I am particularly interested in
how emergent forms of memory and hierarchy allow digital evolution to build programs in
partially-observable and multi-task environments. In addition to general problem solving, my
collaborative research-creation projects apply bio-inspired computing in art/science hybrids
that focus on storytelling, activism, and public engagement.
Email: kellys27 AT msu.edu
Yuan Yuan Postdoc
Yuan Yuan was a Postdoctoral Fellow with the Department of Computer Science and Engineering and a
member of the BEACON Center for the Study of Evolution in Action at Michigan State University,
USA. His research interests include evolutionary computation, machine learning, and search-based
software engineering.
Email: yyuan AT msu.edu
I finished my Ph.D. at Michigan State University in “Evolution of Decision-Making
Systems.” My broad focus is on understanding the cultivation of features that make up
intelligent and robust life-like behavior at all levels of complex systems.
Email: jory AT msu.edu
Honglin Bao (he / him / his) Graduate Student
Honglin was a graduate student at the Banzhaf Lab focusing on the intersection of social
simulation, artificial life, complex systems, and computational social science. More info about
him can be found in his page: carsonhlbao.com
Email: baohongl AT msu.edu
Ken Reid (he / him) Postdoc
Ken's primary research focus was the overlap of evolutionary algorithms with machine learning.
Additionally, he was working on genomic prediction in animals and humans using AI, formulating
problems from the video game `Factorio' for research into real-world optimization problems, and
in employee scheduling problems.
Email: reidken1 AT msu.edu
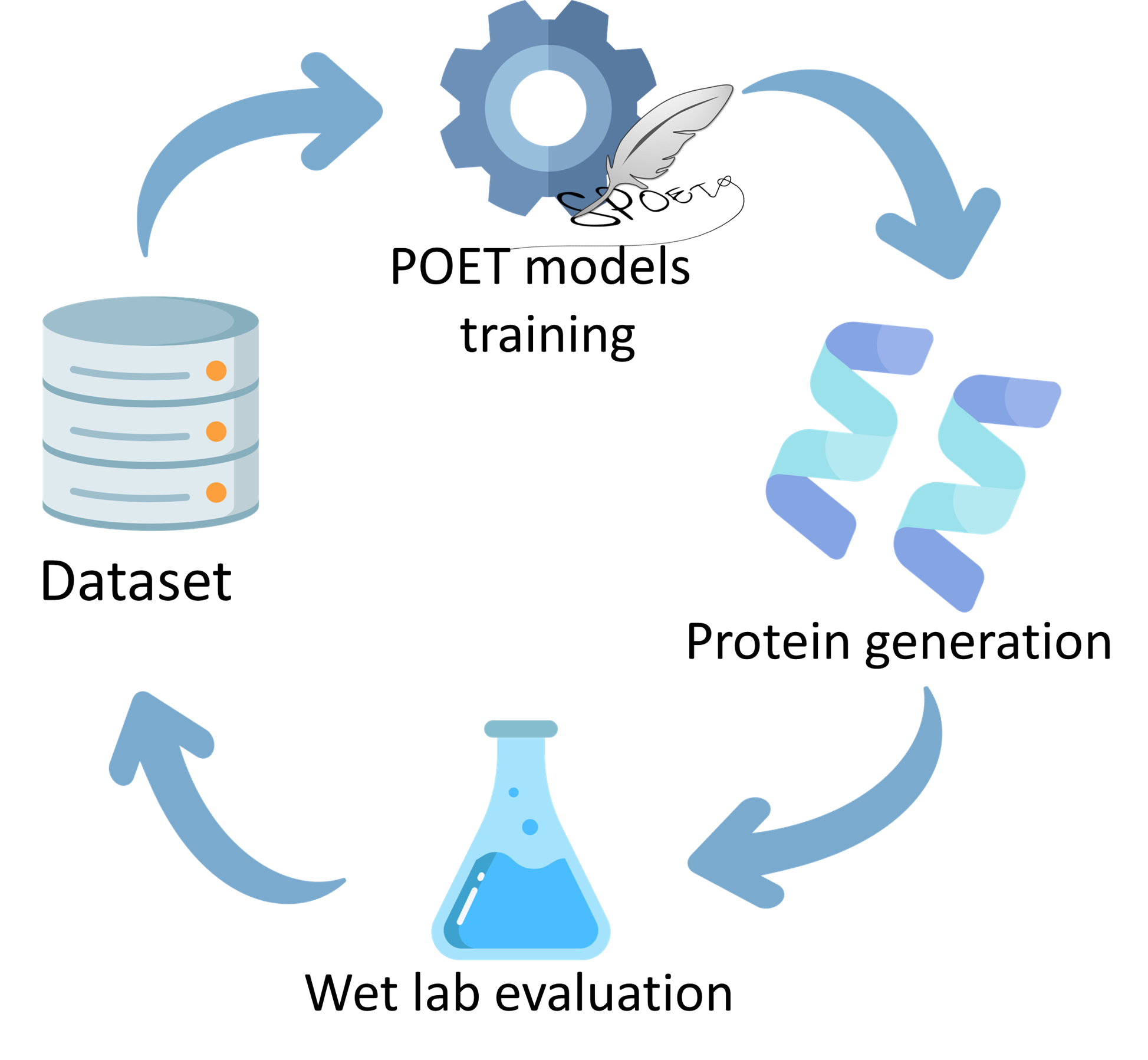
I am currently working on the POET project within the Banzhaf lab, investigating different
techniques to improve automatic peptide design. I received my bachelor's degree in theoretical
physics from Trinity College Dublin and Ph.D. degree in explainable Artificial Intelligence (AI)
(XAI) from the BDS Laboratory, University of Limerick. I was previously a Postdoctoral Research
Fellow within the Complex Software Laboratory, University College Dublin, researching software
testing and mutation analysis and am currently an assistant professor in UCD. My research
interests include grammatical evolution, transfer learning, fuzzy logic, genetic improvement and
XAI.
My thesis subject was about bioinformatics and was focused on genome annotation using deep
learning and genetic programming approaches. I had the opportunity to discover and exploit
bio-inspired algorithms that fascinated me. I joined the Banzhaf lab on September 1st of 2022,
and I am focusing on the development of a new approach based on evolutionary algorithms for the
POET project. I enjoy the fusion of biology with computer science to solve problems.
Email: scalzit1 AT msu.edu
I am CSE PhD student and a researcher at BEACON. I'm interested in bio-inspired algorithms and
applying them on complex real life problems. I'm currently working on my thesis Spatial Genetic
Programming, Automation in Factorio and POET: Protein Optimization Evolving Tool. I love coding
and implementing my ideas specially on computer games!
Email: miralavy AT msu.edu
Jun Guo Graduate Student
I am a PhD student in Mechanical Engineering. I am interested in developing a systematic approach
to generating constitutive models for engineering materials using Genetic Programming. Using our
approach, we can develop a constitutive model with comparable fitness, but it is much simpler
and physically reasonable.
Email: guojun2 AT msu.edu
Salman Ali Graduate Student
I focus on utilizing digital ledger technologies for developing secure, traceable and trust
oriented data sharing applications for food supply chains. My area of research lies in the
overlap of blockchains, machine learning and big data technologies. During my time here, I have
been focussing on using decentralized ledger technologies to collect and process big data for
beef supply chain.
Email: alisalm1 AT msu DOT edu
I am a sophomore undergrad studying Computer Science and Information Science with a Spanish
minor. During my time on this team, I have contributed to research on Multi-Task Reinforcement
Learning and learned about various elements of evolutionary computation through other team
members.
Email: voegerlt AT msu.edu
I am an undergrad student majoring in Computer Science and Mathematics with a minor in
Entrepreneurship and Innovation at Michigan State University. I am working on automatic code
repair and evolutionary program synthesis. I also love spontaneously traveling, listening to
overplayed music and consuming too much coffee.
Email: thammina AT msu.edu
I am a freshman undergrad studying Computer Science with a minor in Computational
Mathematics,
Science, and Engineering. My research interests include machine learning and evolutionary
computation.
Email: wikerale AT msu.edu
I am a junior undergrad studying Computer Science with a minor in Actuarial Science. My research
interests include machine/deep learning and computational finance.
Email: khuctri AT msu.edu
.jpg)


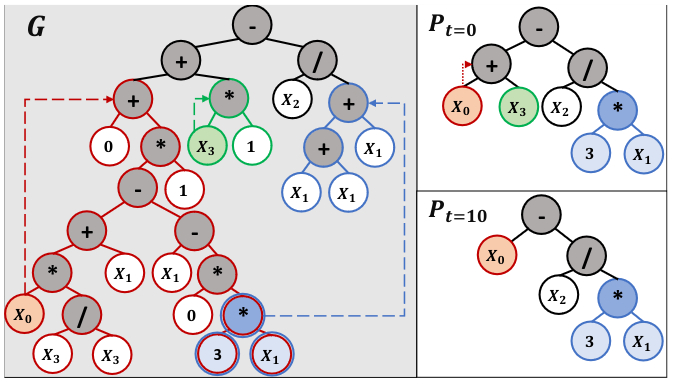
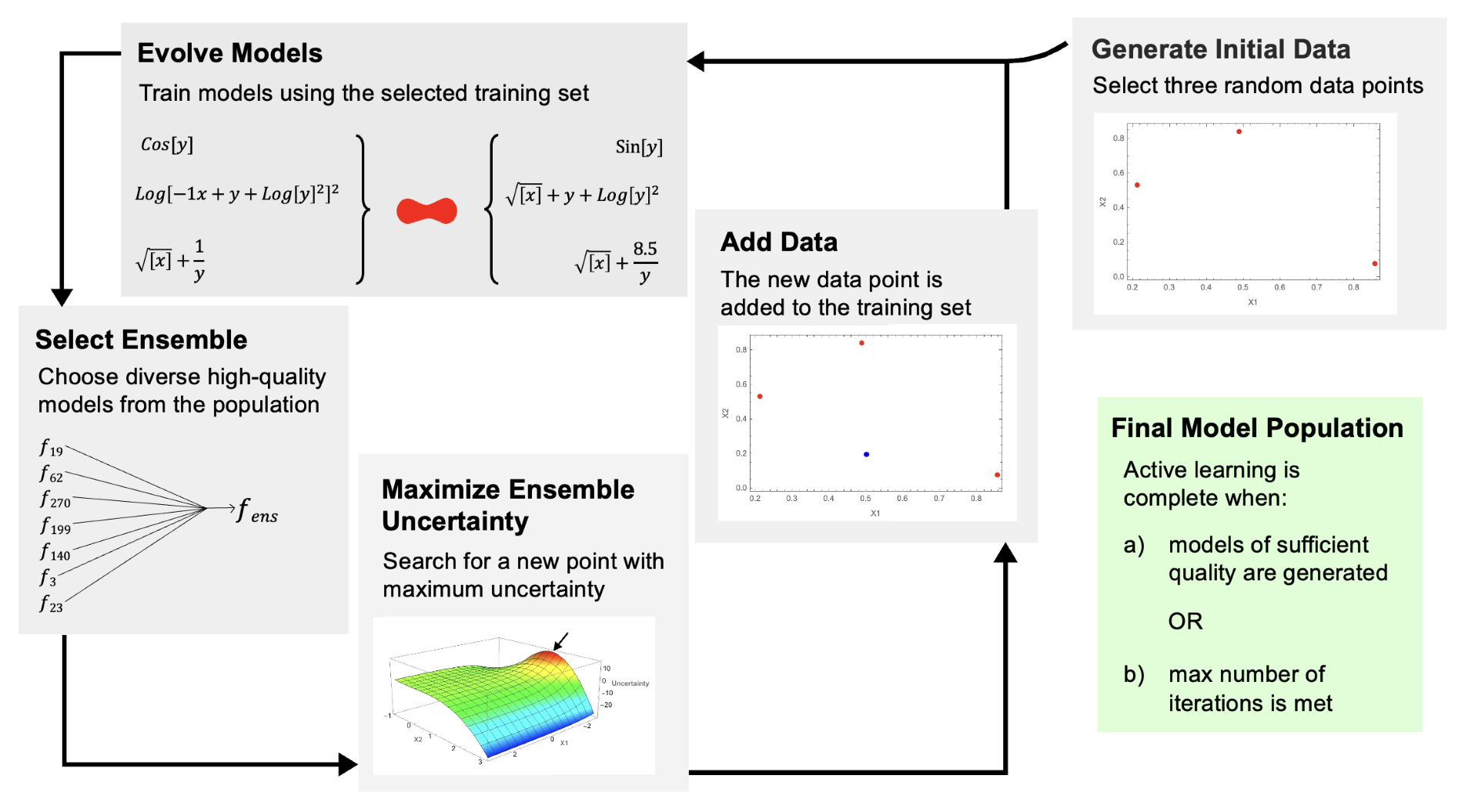

.jpg)





















